Thirteenth paper from a Short-Term Scientific Mission (STSM) (April 21, 2017)
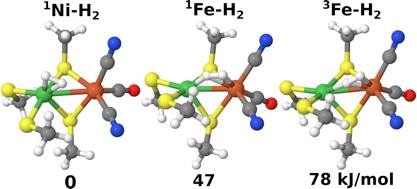
[NiFe] hydrogenases are enzymes that catalyse the reversible conversion of molecular hydrogen to protons and electrons. In this work, the groups of Prof. Pierloot and Ryde report how H2 binds to the active site of this enzyme using computational methods. Combined quantum mechanical and molecular mechanics (QM/MM) optimisation was performed to obtain the geometries. In the QM/MM calculations, TPSS and B3LYP density-functional theory (DFT) methods were employed, and both singlet and triplet state of Ni(II) were considered. To get more accurate energies and obtain a detailed account of the surroundings, the QM/MM calculations with 819 atoms in the QM region were performed. In addition, coupled-cluster calculations with singles, doubles and perturbatively treated triples (CCSD(T)) and cumulant-approximated second-order perturbation theory based on density-matrix renormalisation group complete-active space self-consistent field calculations (DMRG-CASPT2) were carried out using three models to decide what DFT method gives the most accurate structures and energies. The results indicate that H2 binding to Ni in the singlet state is most favourable by at least 47 kJ/mol and that the TPSS functional gives more accurate energies than B3LYP for this system.
The CCSD(T) and DMRG-CASPT2 calculations were performed in the group of Prof. Pierloot in the framework of a STSM by Geng Dong who is a PhD student of Prof. Ryde (COST-STSM-CM1305-30814). This work was recently published in Physical Chemistry Chemical Physics.